Check for model convergence to asymptotic dynamics
Source:R/internal-lin_alg.R, R/utils-export.R
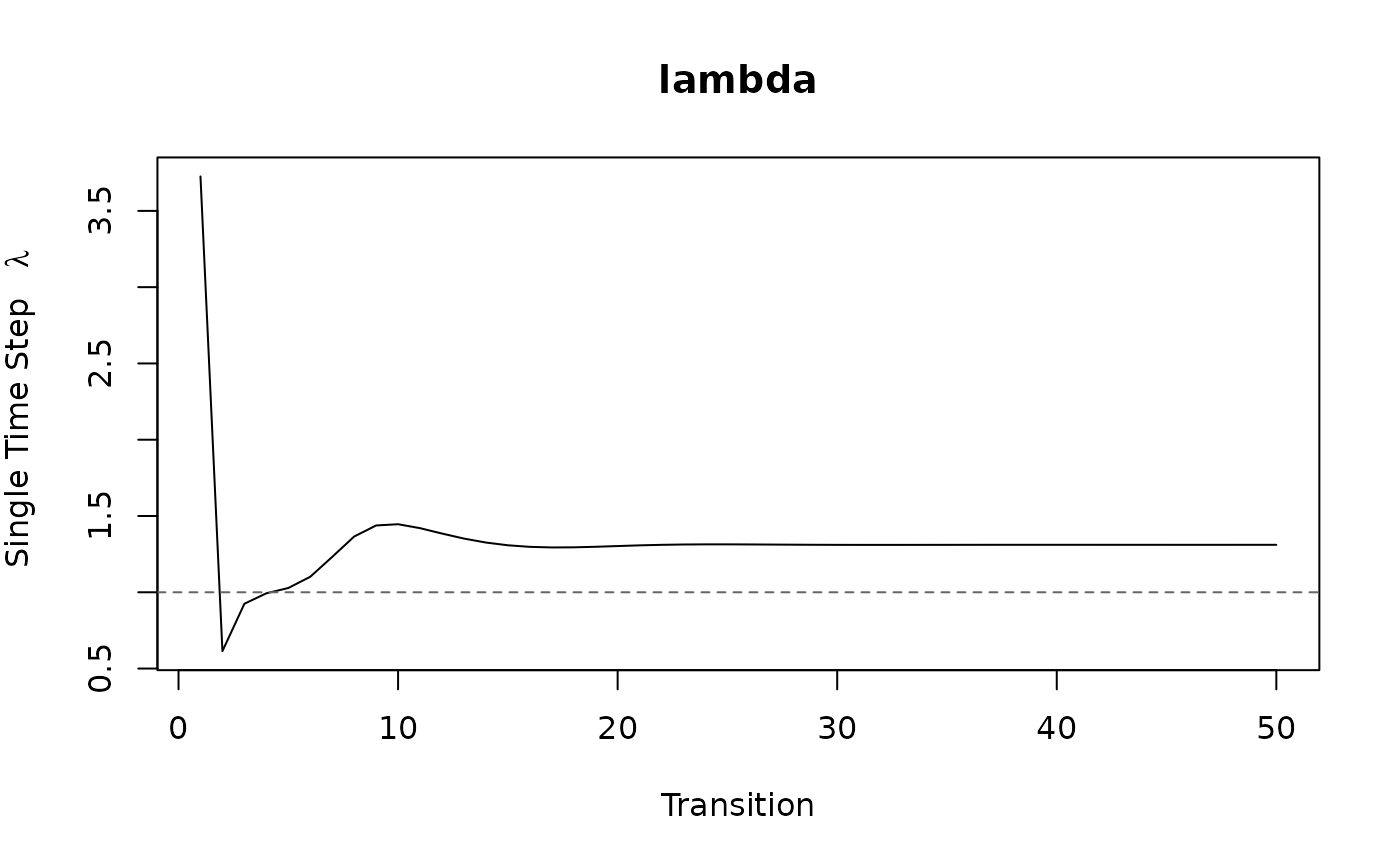
check_convergence.RdChecks for convergence to asymptotic dynamics numerically and
visually. is_conv_to_asymptotic checks whether
lambda[iterations - 1] equals lambda[iterations] within the
specified tolerance, tolerance. conv_plot plots the time series of
lambda (or log(lambda)). For stochastic models, a cumulative mean of
log(lambda) is used to check for convergence.
is_conv_to_asymptotic(ipm, tolerance, burn_in)
# S3 method for class 'ipmr_ipm'
is_conv_to_asymptotic(ipm, tolerance = 1e-06, burn_in = 0.1)
conv_plot(ipm, iterations, log, show_stable, burn_in, ...)
# S3 method for class 'ipmr_ipm'
conv_plot(
ipm,
iterations = NULL,
log = NULL,
show_stable = TRUE,
burn_in = 0.1,
...
)Arguments
- ipm
An object returned by
make_ipm().- tolerance
The tolerance for convergence in lambda or, in the case of stochastic models, the cumulative mean of log(lambda).
- burn_in
The proportion of iterations to discard. Default is 0.1 (i.e. first 10% of iterations in the simulation). Ignored for deterministic models.
- iterations
The range of iterations to plot
lambdafor. The default is every iteration.- log
A logical indicating whether to log transform
lambda. This defaults to TRUE for stochastic models and FALSE for deterministic models.- show_stable
A logical indicating whether or not to draw a line indicating population stability at
lambda = 1.- ...
Further arguments to
plot.
Value
is_conv_to_asymptotic: Either TRUE or FALSE.
conv_plot: codeipm invisibly.
Details
Plotting can be controlled by passing additional graphing parameters
to ....
Examples
data(gen_di_det_ex)
proto <- gen_di_det_ex$proto_ipm %>%
define_pop_state(n_ht = runif(200),
n_b = 200000)
ipm <- make_ipm(proto)
is_conv_to_asymptotic(ipm, tolerance = 1e-5)
#> lambda
#> TRUE
conv_plot(ipm)